This website was created as a project for Genetics 677, an undergraduate course at UW Madison.
Protein Domains
Protein domains are portions of proteins that can evolve separately, develop unique functions, and stand alone from the rest of the protein chain. They often form identifiable three-dimensional structures and can be genetically engineered to transplant into other sequences, forming chimeric proteins. After inserting the amino acid sequence for the human WRN protein, a program called Pfam constructed a linear arrangement of the 5 known protein domains. Clicking on Figure 1 below will link out to an enlarged image of the linear domain sequence.
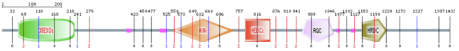
| Figure 1 |
Figure 1. A protein domain representation was created using Pfam, containing 5 protein domains. The green box represents a 3'-5' exonuclease, the red box represents a DEAD/DEAH box helicase, the blue box represents a helicase conserved C-terminal domain, the yellow box represents an RQC domain, and the purple box represents an HRDC domain.
Domains found using Pfam
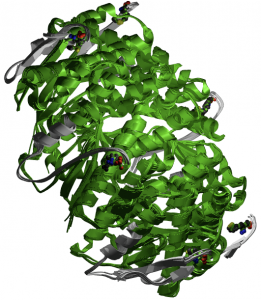
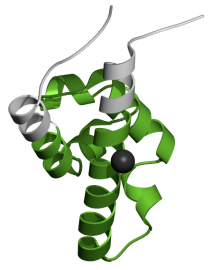
3'-5' Exonuclease. The purpose of this domain is to execute the 3'-5' exonuclease proofreading activity of DNA polymerase I and other enzymes. The domain is 174 amino acids in length, and is found from amino acid 57 to amino acid 230 (2). A three-dimensional representation of the domain can be viewed in Figure 2. The 3'-5' exonuclease domain is conserved among helicases, and is unique within the RecQ family.
Figure 2. Three-dimensional representation of the 3'-5' exonuclease domain created by Pfam.
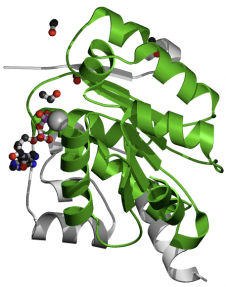
DEAD/DEAH Box Helicase. The purpose of this domain is to unwind DNA for transcription. Additionally, DEAD box helicases are found in processes of RNA metabolism such as pre-mRNA splicing, ribosome biogenesis, nucleocytoplasmic transport, translation, RNA decay and organellar gene expression (3). The domain is 164 amino acids in length and is found from amino acid 550 to amino acid 713. A three-dimensional representation of the domain can be viewed in figure 3.
Figure 3. Three-dimensional representation of the DEAD/DEAH box helicase domain created by Pfam.
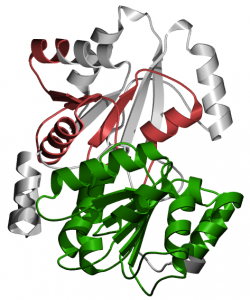
Helicase Conserved C-Terminal Domain. This domain is prevalent in many types of helicases and helicase-related proteins. Although studies are not definitive, this domain may be an integral part of the helicase rather than an autonomously folding unit (4). The domain is 78 amino acids in length and is found from amino acid 782 to amino acid 859. A three-dimensional representation of the domain can be viewed in figure 4.
Figure 4. Three-dimensional representation of the helicase conserved C-terminal domain created by Pfam.
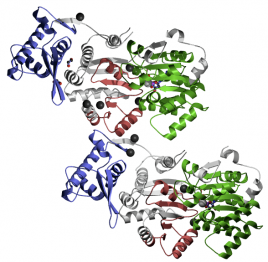
RQC Domain. This sequence contains a helix-turn-helix structure and is a high affinity G4 DNA binding domain (5). The domain is 107 amino acids in length and is found from amino acid 958 to amino acid 1064. A three-dimensional representation of the domain can be viewed in figure 5.
Figure 5. Three-dimensional representation of the RQC domain created by Pfam.
HRDC Domain. This domain is a helicase and RNase D C-terminal that is important for nucleic acid binding (6). The domain is 67 amino acids in length and is found from amino acid 1,153 to amino acid 1,219. A three-dimensional representation of the domain can be viewed in figure 6.
Pfam Analysis: The 5 protein domains found using the Pfam database all had some form of helicase-related activity or DNA-binding theme in common with one another. Helicase needs to bind DNA in order to properly function as a transcriptional enzyme. This is a logical result because the WRN gene codes for a helicase-like protein. It is possible that these protein domains are able to bind to specific DNA sites, affecting transcription. More research needs to be done on these domains to further understand their importance within the WRN gene.
SMART Analysis: Another protein domain database was used to construct a linear arrangement of protein domains within the WRN protein. After inserting the FASTA amino acid sequence for WRN, SMART (Simple Modular Architectural Research Tool) yielded 5 extremely similar domains as Pfam, reinforcing the results of the protein domain search. Clicking on figure 7 below will link out to an enlarged image of the linear domain sequence.
| Figure 7 |
References
1. (Figure 1) Pfam. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk/
2. (Figure 2) Pfam 3'-5' exonuclease domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk/structure/2q3s
3. (Figure 3) Pfam DEAD/DEAH box helicase domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk//family/PF00270.22
4. (Figure 4) Pfam helicase conserved c-terminal domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk//family/PF00271.24
5. (Figure 5) Pfam RQC domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk//family/PF09382.3
6. (Figure 6) Pfam HRDC domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk//family/PF00570.16
7. (Figure 7) SMART. Retrieved March 18, 2010 from http://smart.embl-heidelberg.de/
2. (Figure 2) Pfam 3'-5' exonuclease domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk/structure/2q3s
3. (Figure 3) Pfam DEAD/DEAH box helicase domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk//family/PF00270.22
4. (Figure 4) Pfam helicase conserved c-terminal domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk//family/PF00271.24
5. (Figure 5) Pfam RQC domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk//family/PF09382.3
6. (Figure 6) Pfam HRDC domain. Retrieved March 18, 2010 from http://pfam.sanger.ac.uk//family/PF00570.16
7. (Figure 7) SMART. Retrieved March 18, 2010 from http://smart.embl-heidelberg.de/