This website was created as a project for Genetics 677, an undergraduate course at UW Madison.
Protein Interactions
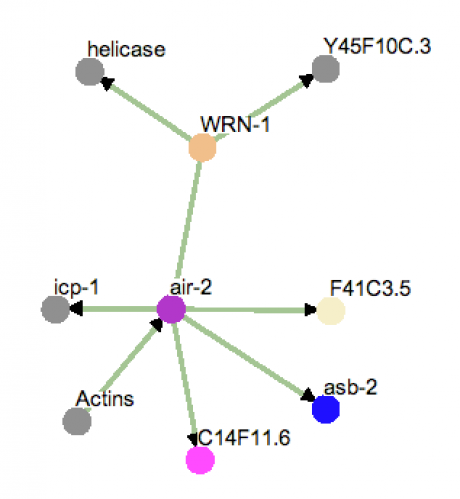
The protein interaction diagram in figure 1 was created using the program OSPREY. By seraching the worm database using the Caenorhabditis elegans orthologue WRN-1, several proteins were identified to share an interaction. Unlike phylogeny trees, the branch lengths in this diagram do not correspond to relatedness, I arranged the tree so the names were readable. Proteins Y45F10C.3, F41C3.5, C14F11.6, Asb-2, icp-1 and helicase are all unsurprising as they either relate to DNA or are protein coding regions.
Analysis: The most notable result is air-2, which is a kinase (phosphorylates proteins) related to the Drosophila aurora and budding yeast IpI1 proteins. This family of kinases has been found in chromosome segregation and mitotic spindle organization. More interestingly, air-2 has recently been identified as a possible target for cancer therapy. Currently there are three Aurora-kinase inhibitors in development, including ZM447439, Hesperadin, and VX-680.
Analysis: The most notable result is air-2, which is a kinase (phosphorylates proteins) related to the Drosophila aurora and budding yeast IpI1 proteins. This family of kinases has been found in chromosome segregation and mitotic spindle organization. More interestingly, air-2 has recently been identified as a possible target for cancer therapy. Currently there are three Aurora-kinase inhibitors in development, including ZM447439, Hesperadin, and VX-680.
Figure 1. WRN homolog WRN-1 protein interaction diagram. The colors correspond to special Gene Ontology (GO) terms that represent each protein. Gray signifies an unknown GO term, blue signifies metabolism, orange signifies DNA metabolism, purple signifies cell organization and biogenesis, pink signifies carbohydrate metabolism, and ivory signifies protein degradation.
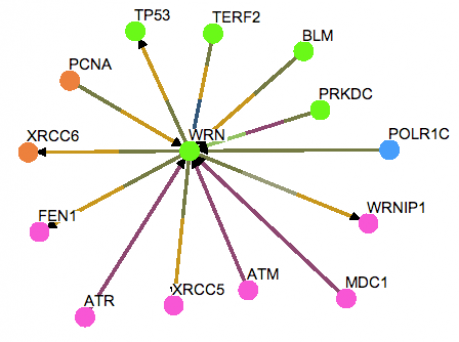
Again using OSPREY, the human database was aligned with WRN. Multiple proteins were identified to share interactions with WRN. As was found in the worm database, many of the protein interactions had a role in DNA replication, and repair.
Analysis: A notable result that is possibly important includes PRKDC, a protein kinase involved in double-stranded DNA break repair and telomere maintenance. This protein may have something in common with air-2, the protein kinase found using the worm database, although a comparison of their protein domains failed to yield any commonality. The other protein of interest found using the human database that was shown to interact with WRN was BLM, the Bloom syndrome protein. It was a little unusual to find BLM in the interaction diagram, although individuals with Bloom syndrome exhibit similar symptoms as those with Werner syndrome. Bloom syndrome can cause cancer and diabetes to develop at extremely early onset, which may suggest that BLM has a more significant relationship with WRN. Comparison of protein domains within BLM and WRN also failed to yield any promising similarity. Therefore, further study of the relationship between BLM and WRN should focus on other genomic and proteomic techniques, such as microarray technologies, to look at mRNA and protein expression levels among individuals with Werner syndrome and Bloom syndrome.
Analysis: A notable result that is possibly important includes PRKDC, a protein kinase involved in double-stranded DNA break repair and telomere maintenance. This protein may have something in common with air-2, the protein kinase found using the worm database, although a comparison of their protein domains failed to yield any commonality. The other protein of interest found using the human database that was shown to interact with WRN was BLM, the Bloom syndrome protein. It was a little unusual to find BLM in the interaction diagram, although individuals with Bloom syndrome exhibit similar symptoms as those with Werner syndrome. Bloom syndrome can cause cancer and diabetes to develop at extremely early onset, which may suggest that BLM has a more significant relationship with WRN. Comparison of protein domains within BLM and WRN also failed to yield any promising similarity. Therefore, further study of the relationship between BLM and WRN should focus on other genomic and proteomic techniques, such as microarray technologies, to look at mRNA and protein expression levels among individuals with Werner syndrome and Bloom syndrome.
Figure 2. WRN human protein interaction diagram. The colors correspond to special Gene Ontology (GO) terms that represent each protein. Green signifies cell organization and biogenesis, orange signifies DNA metabolism, pink signifies DNA repair, and blue signifies a role in transcription.
Wrn from Mus musculus was searched within the mouse database, and LOC569495 from Danio rerio was searched within the Zebra fish database, and neither homolog had any protein interactions using OSPREY.
References
1. (Figure 1 and 2) OSPREY: Network Visualization System. Mount Sinai, NY: Mount Sinai Hospital; 2001. Available from: http://biodata.mshri.on.ca/osprey/servlet/Index