This website was created as a project for Genetics 677, an undergraduate course at UW Madison.
DNA Motifs
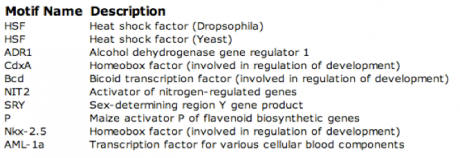
DNA motifs are used in genomics to identify homologous sequences of DNA. This can be useful in identifying functional, structural, or regulatory sequences that may provide further insight into the elements that make up the gene. A program called MOTIF was used to search for DNA motifs. After trying the WRN DNA sequence at cut off scores of 85, 90, 95, and 100, no motifs were found. This is most likely because the entire DNA sequence for WRN is too long for the database to analyze. However, after entering the WRN mRNA sequence into the database, 11 DNA motifs were found using a cut off score of 100. A higher cut off was chosen to reduce the number of motifs and to ensure that they are located in the gene. These motifs and their descriptions can be viewed in figure 1 below.
Figure 1. DNA motifs generated using MOTIF program.
MOTIF Analysis: Several interesting motifs were found using MOTIF, including various Drosophila transcription factors that regulate development, as well as heat shock factors and a plant activator. Heat shock proteins include genes that are expressed when the organism reaches an elevated temperature, or stress response. It is unclear why there were several Drosophila developmental regulatory transcription factors or a plant gene. Perhaps knocking out these motifs would provide greater context into their function within the WRN gene.
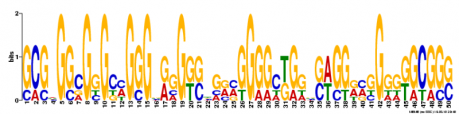
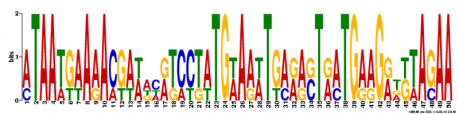
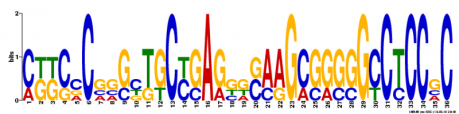
MEME ANALYSIS: Another motif-based sequence analysis tool called MEME was used to search for motifs in WRN. As before with MOTIF analysis, the mRNA sequence for WRN was used as the input. MEME motifs are "represented by position-specific probability matrices that specify the probability of each possible letter appearing at each possible position in an occurrence of the motif." These are displayed below as sequence logos, with the height of an individual letter representing the probability of that nucleotide being present in that specific location multiplied by the total information content of the stack. Although it is an interesting method to visualize the motifs, I am unsure how helpful the sequence logos are for understanding the gene. Three motifs were found and can be viewed in figure 2 below.
Figure 2. Three DNA motifs for WRN created by MEME.
References
1. (Figure 1) MOTIF database. Retrieved March 19, 2010 from http://motif.genome.jp/
2. (Figure 2) MEME database. Retrieved March 19, 2010 from http://meme.sdsc.edu/meme4_4_0/intro.html
3. Timothy L. Bailey and Charles Elkan, "Fitting a mixture model by expectation maximization to discover motifs in biopolymers", Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28-36, AAAI Press, Menlo Park, California, 1994.
4. Kanehisa, M.; Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem Sci. 22, 442-444 (1997).
2. (Figure 2) MEME database. Retrieved March 19, 2010 from http://meme.sdsc.edu/meme4_4_0/intro.html
3. Timothy L. Bailey and Charles Elkan, "Fitting a mixture model by expectation maximization to discover motifs in biopolymers", Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28-36, AAAI Press, Menlo Park, California, 1994.
4. Kanehisa, M.; Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem Sci. 22, 442-444 (1997).