This website was created as a project for Genetics 677, an undergraduate course at UW Madison.
Gene Ontology
Gene ontology is a massive bioinformatic undertaking that attempts to unify gene and gene product components across all species by creating a common vocabulary. Three main goals are the underlying scaffold to the Gene Ontology project; firstly to develop a controlled vocabulary of gene and gene product attributes; secondly, to add commentary to genes and gene products and clearly outline this data; and thirdly, to provide resources to streamline access to all of the data assimilated by the Gene Ontology project. A database called AmiGO was used to search for terms related to WRN. Entries are referred to as Gene Ontology (GO) terms, and a total of 41 GO terms were found in human WRN. To organize the list, GO terms were grouped into 3 categories: biological processes, cellular components, and molecular functions. Lists of the GO terms and their respective graphical representations can be found below. Clicking on each figure will link out to an enlarged image and interactive website.
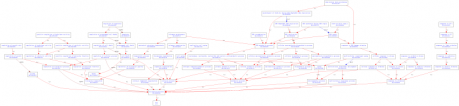
Biological Processes: aging, base-excision repair, cell aging, DNA recombination, DNA synthesis during DNA repair, multicellular organismal aging, positive regulation of hydrolase activity, regulation of apoptosis, regulation of growth rate, replication fork processing, replicative cell aging, response to DNA damage stimulus, response to oxidative stress, response to UV-C, telomere maintenance.
Figure 1. GO terms organized by biological processes created by AmiGO.
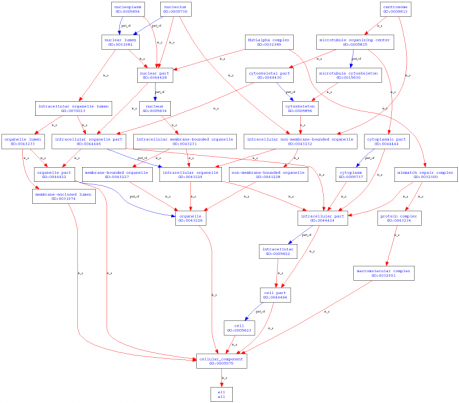
Cellular Components: centrosome, intracellular, MutL alpha complex, nucleolus, nucleoplasm, nucleus.
Figure 2. GO terms organized by cellular components created by AmiGO.
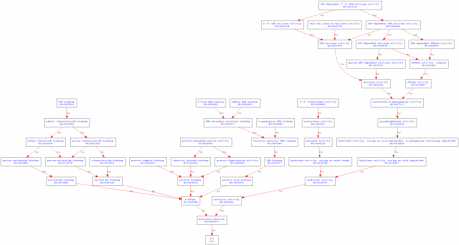
Molecular Functions: 3'-5' DNA helicase activity, 3'-5' exonuclease activity, ATP binding, ATP-dependent 3'-5' DNA helicase activity, ATP-dependent DNA helicase activity, ATPase activity, bubble DNA binding, DNA binding, DNA helicase activity, exonuclease activity, four-way junction helicase activity, G-quadruplex DNA binding, helicase activity, hydrolase activity, nucleotide binding, protein binding, protein complex binding, protein homodimerization activity, Y-form DNA binding.
Figure 3. GO terms organized by molecular function created by AmiGO.
Analysis: All of the GO terms created by AmiGO were unsurprising as they related to DNA binding, cellular aging, and helicase functions. This is logical because WRN is a helicase-like protein. It should be involved in cellular activities that are related to DNA repair, replication, and stress. It also made sense that the cellular component GO terms were all linked to the nucleus, as helicases are localized in the nucleus to perform DNA replication and repair.
References
1. Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, AmiGO Hub, Web Presence Working Group. AmiGO: online access to ontology and annotation data. Bioinformatics. Jan 2009;25(2):288-9.
2. (Figure 1) AmiGO biological processes GO terms for WRN_Human. Retrieved March 19, 2010.
3. (Figure 2) AmiGO cellular components GO terms for WRN_Human. Retrieved March 19, 2010.
4. (Figure 3) AmiGO molecular function GO terms for WRN-Human. Retrieved March 19, 2010.
2. (Figure 1) AmiGO biological processes GO terms for WRN_Human. Retrieved March 19, 2010.
3. (Figure 2) AmiGO cellular components GO terms for WRN_Human. Retrieved March 19, 2010.
4. (Figure 3) AmiGO molecular function GO terms for WRN-Human. Retrieved March 19, 2010.